When Elizabeth Kellogg finished her PhD in 1983, she feared that her skills were already obsolete. Kellogg studied plant morphology and systematics: scrutinizing the dazzling variety of plants’ physical forms to tease out how different species are related. But most of her colleagues had already pivoted to a new approach: molecular biology. “Every job suddenly required molecular techniques,” she says. “It was like I had learned how to make illuminated manuscripts, and then somebody invented the printing press.”
Kellogg had graduated near the start of a revolution in plant biology. Over the next few decades, as researchers adopted molecular tools and DNA sequencing, detailed analyses of plants’ physical traits fell out of fashion. And because many geneticists worked with only a few key organisms, such as the thale cress Arabidopsis thaliana, they didn’t need expertise in comparing and contrasting different plant species. At universities, botany departments folded and molecular-biology departments swelled. Kellogg, now at the Donald Danforth Plant Science Center in St Louis, Missouri, adapted: she embraced genomics, and combined it with her morphology skills to trace the evolution of key traits in the wild relatives of food crops.
But lately, Kellogg has noticed a resurgence of interest in the old ways. Advances in imaging technology — allowing researchers to peer inside plant structures in 3D — mean that biologists are seeking expertise in plant physiology and morphology again. And improvements in gene editing and sequencing have liberated geneticists to tinker with DNA in a wider range of flora, giving them a renewed appetite to understand plant diversity.
Plant biologists hope that, by combining new approaches to botany with data from genomics and imaging labs, they can provide better answers to questions that biologists have asked for more than 100 years: how genes and the environment shape the rich diversity of plants’ physical forms. “People are starting to look beyond their own system into plants as a whole,” says Kellogg. Plant morphology was once a science of form for its own sake, she says, but now, it is being pressed into service to understand how plant traits connect to gene activity across disparate species. “It’s coming back — just under different guises.”
Botany 2.0
Plant morphologists trace their roots back to the eighteenth-century German philosopher and poet Johann Wolfgang von Goethe, who took in the breadth of plant diversity and embarked on a search for an archetypal plant from which all forms could be derived.
A ‘plant archetype’: Johann Wolfgang von Goethe asked French botanist Pierre Jean Turpin to draw this imaginary composite plant, showing off a diversity of forms.Credit: Natural History Museum/Bridgeman
That romantic idea went unfulfilled, but scientists continued his approach of comparing plant structures and functions to learn more about how they evolved and developed. The evolution of flowering plants would later trouble Charles Darwin, who famously called the rapid expansion of such a vast range of flower shapes, colours and pollination strategies an “abominable mystery”.
Although the genomics era led many plant biologists away from morphology, the latest generation of technological advances is steering them back towards the questions that occupied Goethe and Darwin.
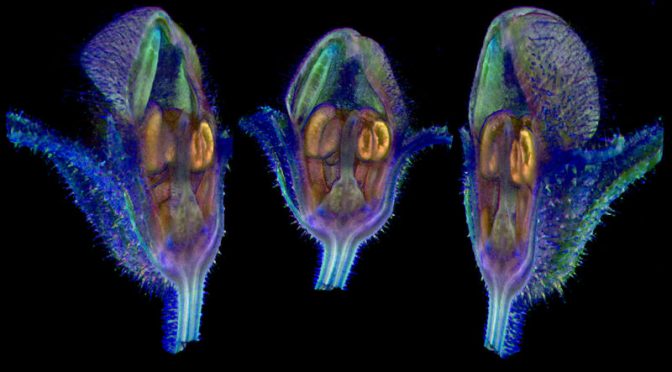
Prominent among these are computed tomography (CT) scanners, which can create 3D reconstructions of internal plant structures without destroying tissue. At the University of Vienna, for instance, plant morphologist Yannick Staedler has used CT scanners to analyse the secrets of a deceptive group of European orchids. Whereas many orchids reward insect pollinators with nectar, others imitate a mating partner or a nectar-rich flower but provide no reward. Biologists back to the time of Darwin have wondered how these ‘deceptive orchids’ thrive, because an insect is unlikely to visit them more than once. Staedler’s studies suggest that such orchids might produce more ovules — the part of the ovary that becomes the seed — potentially to compensate for reduced pollination rates1.
Pollen masses inside a ‘deceptive’ orchid (Orchis militaris), imaged using X-ray computed tomography. Credit: Yannick Staedler
Erika Edwards, a plant morphologist at Yale University in New Haven, Connecticut, is using CT scanners to analyse how the shapes of leaves might be influenced by their early development inside the constrained space of a bud. Botanists have noted for a century that more-serrated, toothed leaves are found in northern, cold regions, whereas smoother leaves are seen in wet tropical forests — but it’s still not clear why. Edwards hopes to unravel the connection.
Some researchers are combining 3D imaging and molecular tools. At the John Innes Centre in Norwich, UK, Enrico Coen’s flower development laboratory uses a technique called optical projection tomography to capture 3D images of plants as they grow. It can also image insect pollinators caught rummaging inside flowers or trapped inside a carnivorous plant. Simultaneously, the group is monitoring gene activity in the plants, by tagging key proteins with fluorescent markers. By combining classical morphology studies with 3D imaging and insights from developmental biology, the group hopes to learn more about the mechanisms that generate plant forms, Coen says. In one study2, for example, he and his collaborators monitored barley-flower development, and explained why that process goes awry in a mutant of barley that was first discovered in the 1830s in Nepal.
Other new imaging techniques are aimed squarely at improving crop breeding. In a field in Jülich, Germany, drones and mini blimps mounted with thermal-imaging cameras fly over plants, while unmanned vehicles called FieldCops carry sensors as they patrol the ground. The effort, at the Jülich Plant Phenotyping Centre, is part of a growing movement to rapidly collect data about plant traits. Initially, these included a limited range of characteristics, such as growth rates or the number of seeds produced. But drones and robots have been fitted with increasingly sophisticated sensors, notes Dirk Inzé, a plant molecular biologist at Ghent University in Belgium. Some are now able to collect data about plant architecture, such as branching and leaf shape, using laser scanners and depth sensors. Similar scanners have been used in lab-grown plants to analyse the rhythmic growth of leaves, and to link that growth to a particular protein complex3.
From genomes to patterns
Molecular labs might also feel a pull back towards botany because, as in other areas of genomics, reading DNA has become so cheap that merely sequencing a plant species is no longer an end unto itself. The first published plant genome — that of A. thaliana — appeared in 2000, and more than 250 plant species have been sequenced since. Now, says William Friedman, director at the Arnold Arboretum of Harvard University in Boston, Massachusetts, “people want to ask how genomes explain evolution and pattern”.
In 2017, for example, the publication announcing the genome of the orchid Apostasia shenzhenica included an analysis of genes that are likely to be responsible for unique aspects of orchid morphology. This includes the labellum, a part of the orchid flower that attracts insects and serves as a landing pad4.
“It’s possible now to understand the paths through which genetic changes influence form,” says Miltos Tsiantis of the Max Planck Institute for Plant Breeding Research in Cologne, Germany. In 2014, his lab used genetics and time-lapse imaging to work out how a particular gene affects leaf shape by restraining cell growth at the leaf’s edge in the mustard species Cardamine hirsuta5. Whereas C. hirsuta’s leaves grow as a series of leaflets around a stem, loss of this gene led to the simple oval leaves found in A. thaliana.
Watching leaves grow: individual cells in this 7-day-old Cardamine hirsuta mustard leaf are outlined by labelling cell membranes with fluorescent molecules.Credit: Miltos Tsiantis/Daniel Kierzkowski
Plant morphologist Dan Chitwood, now at Michigan State University in East Lansing, harnessed sequencing power to look at gene expression in Caulerpa taxifolia — a seaweed that forms complex structures, including a stem and fern-like fronds, from a single, super-sized cell6. Some biologists have argued that the amount and rate of cell division is what shapes plant morphology. But Chitwood’s study showed that gene expression in the unicellular seaweed varies in ways that echo gene expression in similar structures in multicellular plants — suggesting that the dividing cell needn’t always dictate morphology.
Improved molecular tools have now made it possible to tweak DNA in plants that were previously too difficult to work with. The genome-editing tool CRISPR–Cas9 has enabled researchers to tinker with particular genes in a wide range of plants. Researchers have used it to turn purple morning glories white7, for instance, and to alter genes that are involved in building cell walls in orchids8.
But geneticists need to brush up on their botany skills to understand the full implications of these experiments, says Karl Niklas, who studies plant evolution at Cornell University in Ithaca, New York. Researchers often knock genes out to determine how they affect a plant’s form or function. “If you’re not really capable of diagnosing the morphology or the anatomy, you really don’t know what you’re looking at,” Niklas says.
He recalls a time when a student came to him with a mutant form of maize (corn) to show how the xylem — the collection of tubes that carry water and nutrients from the roots to the rest of the plant — was deformed. But the student was actually looking at normal phloem, a different network of vessels with a distinct structure that distributes nutrients formed in the leaves. “You know, it just makes your teeth hurt,” he says.
Researchers also lose out when they do not take the time to consider the diversity of plant forms in nature, says Chelsea Specht, a plant biologist also at Cornell University. She has seen cases in which scientists have failed to realize that their genetic mutants — for instance, Arabidopsis mutants with altered branching patterns — are recapitulating naturally occurring plant forms found in other lineages. When this happens, she says, researchers miss opportunities to put traits into an evolutionary context.
Botany bootcamps
The prospect of fading expertise so worried Friedman that, in 2013, he and his wife, plant morphologist Pamela Diggle of the University of Connecticut in Storrs, launched an intensive botany bootcamp for biologists. “It’s been one of my missions as an academic to keep that knowledge going,” says Diggle. “It’s important to keep this information alive in the community.”
The programme was first funded by the US National Science Foundation, and the New Phytologist Trust, a plant-science non-profit organization in Lancaster, UK, plans to pick up the bill from this year. It accepts about a dozen scientists each year, some from laboratories that typically focus on molecular biology and genomics. The course routinely has about six times as many applicants as positions, says Friedman.
Jamie Kostyun, an evolutionary geneticist, took the course in 2013 to gain the skills she needed to explore the floral traits of the genus Jaltomata. These species are kin to kitchen staples such as tomatoes and potatoes, but they boast a remarkable and recently evolved diversity of flowers. Some are flat, others tubular; some reward pollinators with sticky orange nectar, others ooze a blood-red sweet treat.
“They have crazy floral variation that nobody has looked at before,” Kostyun says. “I wanted to understand where that diversity came from.” She has used her plant-morphology training to detail the development of flowers in five Jaltomata species in her PhD thesis. Now, as a postdoc at the University of Vermont in Burlington, she is studying the panoply of nectar compositions and genetically analysing the vast array of flower shapes.
Friedman hopes that others will follow in Kostyun’s footsteps, uniting these approaches with classical comparative techniques and generating insights into questions that have dogged researchers for decades. “What did the first flowers look like? You could probably open a book from 1900 and still ask the same questions that people were asking about basic plant structure,” he says. “We know more now, but we don’t necessarily know the answers.”